South China Agricultural University College of Horticulture released a high-quality chromosome-level genome map of dragon fruit
Pitaya is native to tropical deserts in Central America. It is a perennial climbing herbaceous fruit tree of the Caryophyllales (Cactaceae) family of Cactaceae (Hylocereus) or Selenicereus (Selenicereus). It has the advantages of long, high yield, no large and small years, and easy cultivation and management. Pitaya fruit is rich in nutrition and unique in function. It contains plant albumin, water-soluble dietary fiber and betain which are rare in general plants. It has the effects of detoxification, nourishing lungs, improving eyesight, weight loss, lowering blood sugar, and moisturizing intestines. The seeds of dragon fruit contain various enzymes, unsaturated fatty acids and antioxidant substances, which have the functions of anti-oxidation, anti-free radicals, anti-aging, and whitening skin. Dragon fruit flowers and stems can be used as vegetables or medicines, and the peel can be used to extract pigments for food additives. In addition, dragon fruit has a long flowering period (April to November), the flowers are large and fragrant and beautiful, and the fruit is large and beautiful. It is suitable for potted plants, bonsai or garden cultivation and viewing, and can also be used to build sightseeing picking orchards. It can be seen that dragon fruit integrates fruit, flowers, vegetables, health care and medicine. It has high economic, nutritional and medicinal value, and is loved by consumers and growers. At present, dragon fruit breeding is still in the traditional breeding stage, and molecular breeding technology is still in the basic research stage; the biological mechanism of quality formation and maintenance is also limited. One of the main reasons is the lack of high-quality dragon fruit genome maps.
Recently, Horticulture Research published an online research paper entitled A chromosome-scale genome sequence of pitaya (Hylocereus undatus) provides novel insights into the genome evolution and regulation of betalain biosynthesis by the College of Horticulture, South China Agricultural University, deciphering the genome of red skin and white flesh dragon fruit , Reached the level of high-quality chromosome assembly, constructed a high-density genetic map of dragon fruit, screened 43 structural genes involved in dragon fruit betanin biosynthesis and 557 transcription factors that may regulate them.
The research used the sequencing technology of PacBio, Illumina, 10× Genomics and Hi-C to sequence and assemble the genome of the red-skinned and white-fleshed dragon fruit cultivar ‘GHB’ (‘Guanhuabai’, H.undatus) (Figure 1). The assembled size is 1,386.95 Mb, the scaffold number is 675, the N50 length is 127.15 Mb, the contigs number is 7647, and the N50 length is 0.58 Mb. Through Hi-C technology, 97.67% of the sequence can be anchored to 11 dragon fruit chromosomes. The chromosome length ranges from 97.47 Mb to 146.67 Mb, and the GC content is 36.9%. The BUSCO and CEGMA indexes are 93.8% and 93.55% respectively. The comparison rate and coverage rate of BWA were 99.68% and 95.20%, respectively. The results show that the ‘GHB’ genome data has high integrity, continuity and assembly quality at the chromosome level.
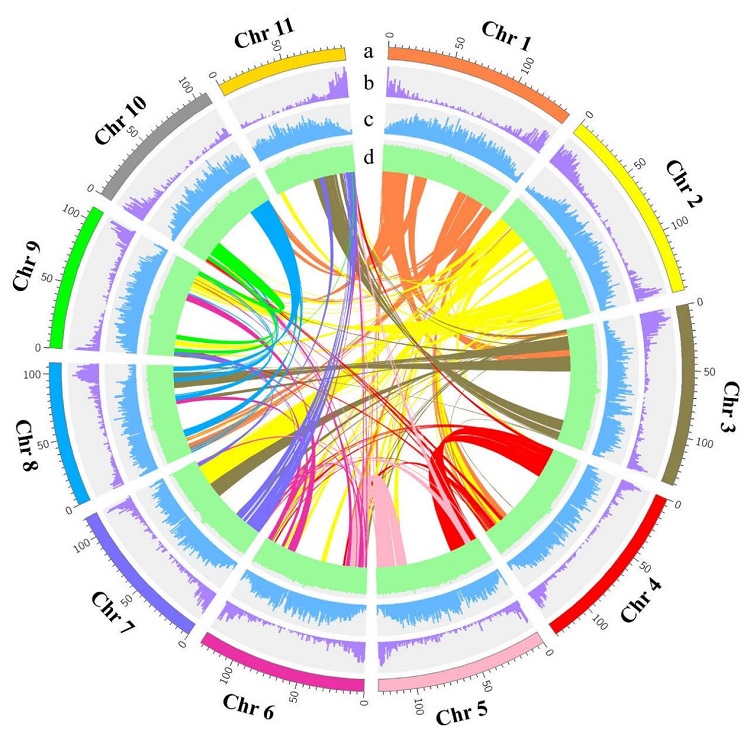
Figure 1 Genome characteristics of dragon fruit. a. Chromosome; the unit is Mb; b. gene density; c. tandem repeat density; d. GC content; the middle region is genome collinearity.
The dragon fruit genome was further predicted and annotated through de novo prediction, RNA-Seq and homology search, and finally 27,753 CDS, 4,989 miRNAs, 4,857 tRNAs, 5,909 rRNAs and 3,877 snRNAs were obtained. Through comparative genomics (11 species including dragon fruit), 13,594 gene families, 517 dragon fruit-specific gene families, and 419 single-copy homologous gene families were identified. According to the results of collinearity between dragon fruit and spinach genomes, as well as the 4dTv analysis results of beet and spinach genomes, dragon fruit, beet, and spinach had the earliest WGT event (Dianthus was isolated from Arabidopsis thaliana), followed by a recent event. The WGD event (Pitaya is separated from Dianthus). Using 419 single-copy homologous gene families to construct a phylogenetic tree, it is inferred that the separation time of Dianthus from Arabidopsis is about 118.2 Mya, and the separation time of Pitaya from Dianthus is about 65.3 Mya.
Betanin is an important plant secondary metabolite similar to anthocyanin. It not only plays a role in coloring in plants, but also plays an important role in the process of plants adapting to adversity. The skin and flesh of dragon fruit show different colors, which are mainly derived from the synthesis and accumulation of betanin. In this thesis, 203'GHB'×'DaHong' (red skin and red flesh dragon fruit) F1 hybrid populations were used as materials to construct a high-density dragon fruit genetic map, and 20,872 bin markers (56,380 SNPs) were obtained, which can be used for subsequent QTL analysis . Using'GHB' and'GHH' (red skin and red pitaya) pitaya as materials for transcriptome sequencing, 43 structural genes involved in the biosynthesis of pitaya betanin were screened, including 3 ADHs, 11 CYP76AD1s, and 5 One TYRs, six DDCs, three DODAs, two cDOPA5GTs, eight B5GTs, and five B6GTs (Figure 2). WGCNA analysis of differentially expressed genes (DEGs) found that they were enriched into 23 modules. The expression trends of 145, 112, 825 genes in the three modules lightcyan (0.73), grey60 (0.72), blue (0.69) and betanin There is a high correlation coefficient in the content change of fruit growth and development; 557 transcription factors and 43 structural genes are co-expressed, of which MYB, bHLH, AP2-EREBP and HB gene families account for the largest proportion.
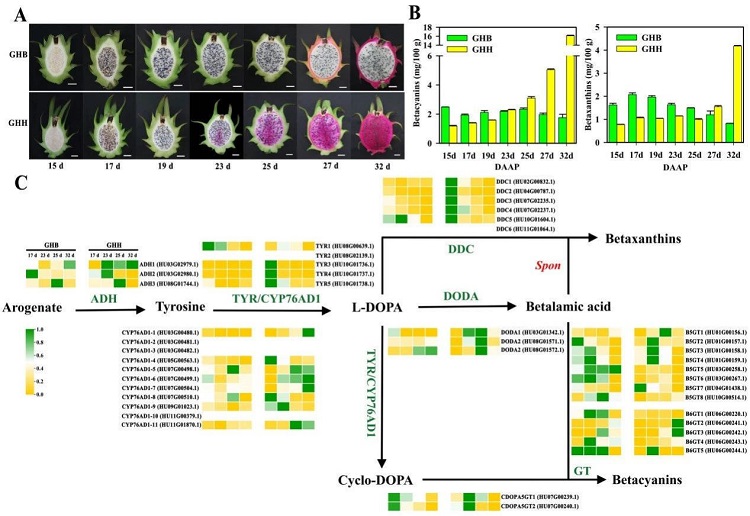
Figure 2 Pitaya betain biosynthesis pathway. A. ‘GHB’ and ‘GHH’ pitaya pulp; B. Betatin content in ‘GHB’ and ‘GHH’ pitaya pulp; C. Expression level of structural genes in the betatin biosynthesis pathway.
In summary, this study used PacBio, Illumina, 10× Genomics and Hi-C sequencing technology to perform De nove sequencing on the red skin and white flesh dragon fruit genome, and obtained a high-quality chromosome-level genome map. The genetic research and molecular breeding provide an important theoretical basis.
Professor Chen Jianye, Dr. Xie Fangfang and Nuohe Zhiyuan Cui Yanze from the College of Horticulture, South China Agricultural University are the co-first authors of the paper. Professor Qin Yonghua and Professor Hu Guibing from the College of Horticulture, South China Agricultural University and Researcher Sun Qingming from the Institute of Fruit Research, Guangdong Academy of Agricultural Sciences are the correspondence of the paper. author. This project was funded by the National Natural Science Foundation of China (31972367 and 31960578), the Guangzhou Scientific Research Program Key Project (201904020015), and the Guangdong Province Key Field Research and Development Program (2018B020202011).
Article link: https://www.nature.com/articles/s41438-021-00612-0